Neutrophils and galectin-3 defend mice from lethal bacterial infection and humans from acute respiratory failure - Nature.com
Abstract
Respiratory infection by Pseudomonas aeruginosa, common in hospitalized immunocompromised and immunocompetent ventilated patients, can be life-threatening because of antibiotic resistance. This raises the question of whether the host's immune system can be educated to combat this bacterium. Here we show that prior exposure to a single low dose of lipopolysaccharide (LPS) protects mice from a lethal infection by P. aeruginosa. LPS exposure trained the innate immune system by promoting expansion of neutrophil and interstitial macrophage populations distinguishable from other immune cells with enrichment of gene sets for phagocytosis- and cell-killing-associated genes. The cell-killing gene set in the neutrophil population uniquely expressed Lgals3, which encodes the multifunctional antibacterial protein, galectin-3. Intravital imaging for bacterial phagocytosis, assessment of bacterial killing and neutrophil-associated galectin-3 protein levels together with use of galectin-3-deficient mice collectively highlight neutrophils and galectin-3 as central players in LPS-mediated protection. Patients with acute respiratory failure revealed significantly higher galectin-3 levels in endotracheal aspirates (ETAs) of survivors compared to non-survivors, galectin-3 levels strongly correlating with a neutrophil signature in the ETAs and a prognostically favorable hypoinflammatory plasma biomarker subphenotype. Taken together, our study provides impetus for harnessing the potential of galectin-3-expressing neutrophils to protect from lethal infections and respiratory failure.
Similar content being viewed by others
NLRP6 modulates neutrophil homeostasis in bacterial pneumonia-derived sepsis

Locally organised and activated Fth1hi neutrophils aggravate inflammation of acute lung injury in an IL-10-dependent manner
G-CSFR antagonism reduces neutrophilic inflammation during pneumococcal and influenza respiratory infections without compromising clearance
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium that can be cleared by host defense mechanisms from the respiratory tract of healthy individuals but can cause pneumonia with a high degree of morbidity and mortality in immunocompromised patients1,2,3. P. aeruginosa is also a leading cause of acute infections resulting in ventilator-associated pneumonia (VAP) in hospitalized immunocompetent patients4 and can cause chronic infections in cystic fibrosis patients5. Infections by P. aeruginosa can be life-threatening and difficult to treat because the bacterium can acquire wide range antibiotic resistance through various genetic mechanisms6. Therefore, it is important to consider alternate avenues to fight this pathogen, such as by harnessing the host's immune response.
Bacterial lipopolysaccharide (LPS), present in the outer membrane of Gram-negative bacteria, is an immunomodulatory agent7. An early study showed the ability of LPS to induce resistance to subsequent infections by different Gram-negative bacteria8. LPS-induced resistance results in reduced bacterial burden with promotion of leukocyte recruitment9. These findings suggest that LPS can equip the immune system to better defend the host against the invading pathogen. However, the mechanisms underlying the protective effects of LPS are not well understood. Here we show that prior exposure to a low dose of LPS protects mice from a lethal infection by P. aeruginosa. Pre-exposure to LPS in PA-infected mice causes accumulation of a neutrophil population uniquely enriched in both phagocytosis-associated genes and a cell-killing gene set comprising Lgals3, which encodes the antibacterial protein, galectin-3. Functional studies in mice support the impact of LPS on neutrophils in conferring protection. In humans, survivors of acute respiratory failure show significantly higher galectin-3 levels in endotracheal aspirates (ETAs) compared to non-survivors, galectin-3 levels strongly correlating with a neutrophil signature in the ETAs and a prognostically favorable hypoinflammatory plasma biomarker subphenotype.
Results
Pre-exposure to LPS protects mice from a lethal infection by P. aeruginosa with signature of a regulated inflammatory response
We sought to determine whether LPS can protect mice from a lethal infection by PA14, a strain commonly used to study P. aeruginosa infection of mice10,11. Mice intratracheally (i.t.) infected with 106 colony forming units (cfu) of PA14 reproducibly appeared huddled and ill by 4–6 h after infection. We first assessed bacterial load in the lung tissue and in the peripheral blood by infecting mice on day 1 or 3 after i.t. administration of a low dose of 20 μg of ultrapure LPS. All mice were sacrificed at 4 h post-infection, a time point at which infected mice that were not pre-exposed to LPS appeared visibly ill (Fig. 1a). A significantly lower bacterial load was detected in the lungs of mice after exposure to LPS compared to that in mice that did not receive any LPS (Fig. 1b). No dissemination of bacteria in the blood was observed in LPS-treated mice whether infected on day 1 or 3 after LPS exposure, which was clearly evident in the control infected mice, all mice sacrificed at 4 h after infection (Fig. 1c). The mice infected with PA14 without LPS pre-exposure appeared near death after 18–20 h and were euthanized (Fig. 1d). This time point was considered the time of death for mice not pre-exposed to LPS since our Institutional Animal Care and Use Committee (IACUC) rules do not permit death as an end-point in survival studies. In contrast, all PA14-infected mice that received LPS 3 days earlier behaved normally without displaying any evidence of sickness and this was followed up to 14 days after infection (Fig. 1d). LPS-mediated protection was observed in both sexes. Thus, these experiments established 106 cfu as a lethal dose of infection and the two groups of mice with or without LPS pre-exposure are hereafter referred to as LPS + PA14 and PA14 respectively. Bronchoalveolar lavage fluid (BALF) albumin concentration, indicative of alveolar permeability, was significantly higher in the PA14 mice compared to that in the LPS ± PA14 mice (Fig. 1e).
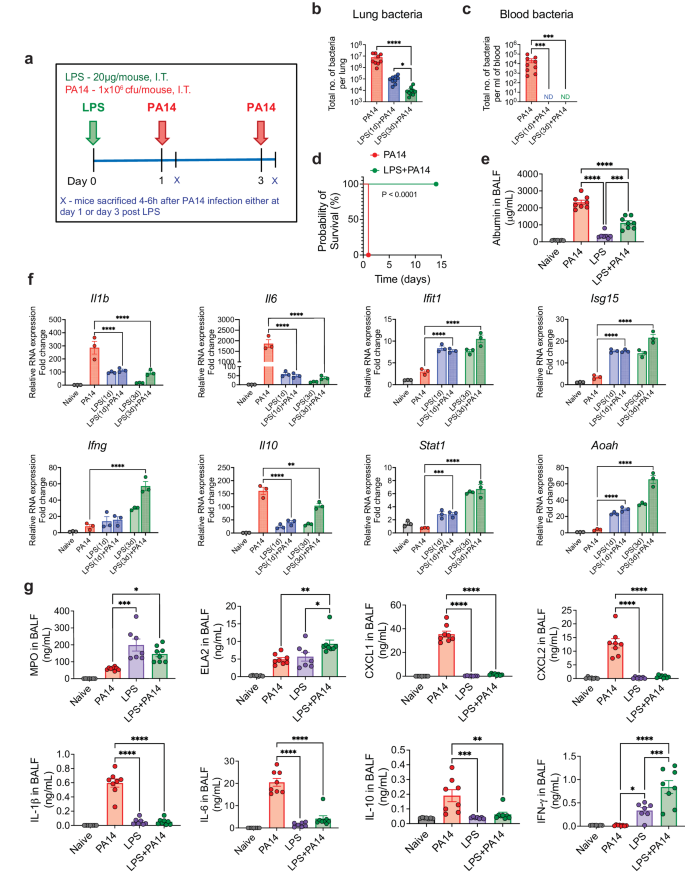
a Schematic of PA14 infection with or without LPS pre-treatment. Bacterial load showing reduced bacterial burden in the lung (b) and decreased peripheral dissemination (c) in LPS + PA14 mice compared to PA14 mice (4 h post-infection). n = 9 (PA14), 9 (LPS(1d)+PA14), and 10 (LPS(3d)+PA14) mice. Data were log transformed using log base 10 to adjust for differences in standard deviation prior to analysis. Data pooled from 3 independent experiments and analyzed using one-way ANOVA with the Dunn's test. d Kaplan-Meier survival curve showing 100% mortality of PA14 infected mice within 18–20 h of infection, whereas 100% survival of LPS + PA14 group as monitored up to 14 days post infection. n = 8 (PA14 and LPS + PA14) mice per group. Data representative of 3 independent experiments and analyzed using Log-rank test. e Higher albumin levels in BALF of PA14 mice compared to LPS ± PA14 mice (mice pre-exposed to LPS for 3 days), BALF being collected 4 h post-infection from both groups of mice and also from naïve mice and mice only treated with LPS. n = 7 (Naïve and LPS), and 8 (PA14 and LPS + PA14) mice. Data pooled from 2 independent experiments and analyzed using ordinary one-way ANOVA with the Tukey post-hoc test. f RT-qPCR analysis of gene expression of Il1b, Il6, Ifit1, Isg15, Ifng, Il10, Stat1 and Aoah in the lung tissue of the four groups of mice. n = 3 mice per group. Data representative of 3 independent experiments and analyzed using ordinary one-way ANOVA with the Dunnett's test. g MPO and ELA2 protein levels associated with neutrophil influx and chemokine and cytokine levels in the BALF of the two groups of mice. n = 7 (Naïve and LPS), and 8 (PA14 and LPS + PA14) mice. Data pooled from 2 independent experiments and analyzed using ordinary one-way ANOVA with the Tukey post-hoc test. All data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data and exact P values are provided as a Source Data file.
We measured mRNA levels of key genes associated with host defense by RT-qPCR and also assayed cytokine and chemokine levels along with markers of neutrophil accumulation, critical for defense against PA12, in the lung. The RT-qPCR data revealed differential expression of infection-response genes between the LPS + PA14 and PA14 mice. The PA14 mice mounted a significantly greater expression of the cytokine genes (Il1b, Il6) compared to the LPS + PA14 mice (Fig. 1f). In contrast, the expression of the interferon stimulated genes (ISGs), Ifit1 and Isg15, and Ifng were significantly higher in the LPS + PA14 mice compared to that in the PA14 mice. The expression of the anti-inflammatory cytokine, Il10, was attenuated in the LPS + PA14 mice, albeit expressed above baseline (Fig. 1f). We also examined the expression of Stat1, a transcription factor activated by both Type I and II interferons (IFNs), which was higher in the LPS + PA14 mice compared to that in the PA14 mice (Fig. 1f). The profile of gene expression in the LPS-treated mice prompted us to also assess the expression of the enzyme Acyloxyacyl hydrolase (Aoah) that specifically facilitates the removal of secondary fatty acyl chains from the lipid A moiety of LPS thereby limiting the toxic effects of LPS and yet allowing an immune response after LPS exposure13,14,15. Aoah expression was low in both naïve and PA14-infected mouse lungs but its expression was significantly higher in response to LPS, with highest expression in the LPS + PA14 mice on day 3 post-LPS exposure (Fig. 1f) when the lung bacterial load was the lowest (Fig. 1b). We assayed mediators present in the BALF collected on day 3 after LPS or phosphate-buffered saline (PBS) exposure followed by PA14 infection. Significantly greater levels of myeloperoxidase (MPO) and neutrophil elastase (ELA2), both associated with neutrophil influx, were detected in the LPS + PA14 mice compared to that in the PA14 mice (Fig. 1g). In contrast, while the levels of the neutrophil chemoattractants, CXCL1 and CXCL2, in the BALF were significantly higher above baseline in the PA14 mice within 4 h after infection, this response was attenuated in mice pre-exposed to LPS, again showing a fine tuning of the inflammatory response by LPS (Fig. 1g). In agreement with the RT-qPCR data, higher levels of the cytokines IL-1β, IL-6 and IL-10 were detected in the BALF of mice infected with PA14 alone compared to those pre-exposed to LPS and then infected (Fig. 1g). However, IFN-γ protein level was higher in the BALF of the LPS + PA14 mice compared to that in the PA14 mice, also consistent with the RT-qPCR data (Fig. 1g). Histological assessment of the lung sections of the LPS + PA14 mice revealed that an expected inflammatory response was detectable in the lungs on day 3 but the inflammation resolved 3 days later (Supplementary Fig. 1a) consistent with the normal behavior of these mice followed up to 14 days after PA14 infection (Fig. 1d).
Increase in neutrophils and interstitial macrophages in LPS-exposed lungs
We analyzed single cell suspensions of lung tissue from PA14 and LPS ± PA14 mice by multi-color flow cytometry using an optimized gating strategy (Supplementary Fig. 1b). In these and subsequent experiments, we focused on the host response to infection on day 3 after LPS treatment when the lung bacterial load was the least as compared to that in mice without LPS pre-exposure (Fig. 1b). In the 3 groups of mice-PA14, LPS and LPS + PA14, total lung cell counts significantly increased compared to that in naïve mice with a similar profile observed for CD11b+ cells (Fig. 2a). However, we detected a loss of SiglecF+CD11blo/− alveolar macrophages (AMs) in these groups from numbers present in naïve mice (Fig. 2a, b). Loss of AMs has been reported in many settings of lung infection and is referred to as the macrophage disappearing reaction16,17,18. Compared to PA14 mice, lungs of both LPS only and LPS + PA14 mice showed significantly higher numbers of CD11b+Ly6G+ neutrophils and CD11b+CD64+CD24− interstitial macrophages (IMs) were detected, both cell types having high phagocytic functions. Neutrophil numbers were only slightly higher in the LPS + PA14 mice compared to that in LPS only mice (Fig. 2a). This profile also matched the lung histology profiles (Supplementary Fig. 1a). In contrast, the numbers of CD11b+Ly6ChiCD43− classical monocytes (cMos), did not appreciably increase in response to either LPS or PA14 (Fig. 2a, b). The flow cytometry data also revealed significantly lower numbers of IMs in the PA14 mice as compared to that in the LPS-exposed mice (Fig. 2a, b). Taken together, our data suggested that although a robust neutrophilic response was mounted in both the LPS + PA14 and PA14 mice, given the divergence in the outcome in the two groups, functionally the neutrophils in the LPS + PA14 mice were more adept in bacterial clearance compared to those in the PA14 mice (Fig. 1b). Another feature of LPS pre-exposure was the significant increase in Ly6G+CD14+IFN-γ+ neutrophils as compared to a very small percentage detected in the PA14 group (Fig. 2c).
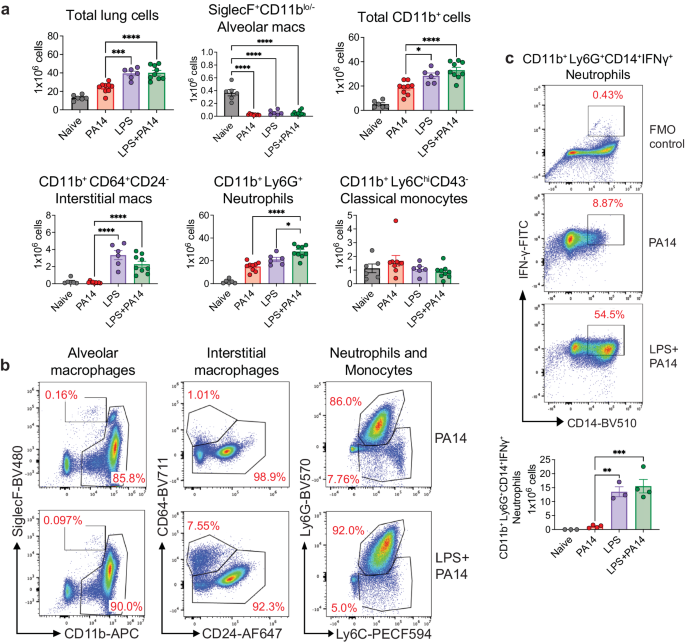
a Flow cytometry analysis showing total numbers of lung cells, AMs, CD11b+ cells, neutrophils, classical monocytes, and IMs. n = 6 (Naïve and LPS), and 9 (PA14 and LPS + PA14) mice. Data pooled from 2 independent experiments and analyzed using ordinary one-way ANOVA with the Tukey post-hoc test. b Representative flow plots for identification of the cell types in the lungs of the two groups of mice. c Representative flow plots and total counts of IFN-γ-expressing neutrophils in the lungs of the two groups of mice from 2 independent experiments. n = 3 (Naïve and LPS), and 4 (PA14 and LPS + PA14) mice. Data analyzed using ordinary one-way ANOVA with the Tukey post-hoc test. All data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data and exact P values are provided as a Source Data file.
Pre-exposure to LPS in PA-infected mice promotes specific neutrophil and IM populations as revealed by scRNA-seq
Next, for an in-depth analysis of the immune cells in the LPS + PA14 and PA14 groups, we took the approach of single cell RNA-sequencing (scRNA-seq). As performed for flow cytometry analysis (Fig. 2a–c and Supplementary Fig. 1b), mice received i.t. LPS or PBS, followed in 3 days with PA14 via the same route. Mice were sacrificed at 4 h post infection to harvest CD11b+ immune cells from the lungs for scRNA-seq analysis. We focused on CD11b+ cells based on our flow cytometry data that showed depletion of CD11blo/− AMs, with concomitant increase in neutrophils and IMs, these cell types and other innate immune cells being largely CD11b+. Here, we implemented the Seurat based integrative data analysis pipeline to analyze the single cell data from LPS + PA14 and PA14 samples. After additional quality filtering, we profiled 46,826 cells from LPS + PA14 and PA14 samples. We identified 14 discrete cell types based on distinct markers and after merging similar clusters noted for IM2 (4 and 5), N3 (7 and 8) and N4 (0 and 1) (Fig. 3a, b and Supplementary Fig. 2a–c). The three biological replicates from the same condition (i.e., PA14 vs. LPS + PA14) were merged using the Seurat function merge where each condition is in the form of a combined data matrix but we can still identify which replicate the cells come from. UMAP and tSNE plots are standard representations to depict how well the replicates overlap each other (Supplementary Fig. 3). Manually curated annotations were further substantiated with automated annotations drawn from Immune Genome (ImmGen) database19 using the SingleR package. The major cell clusters identified included four neutrophil populations, three IM populations, monocytes (Ly6C+Lyve1lo), mixed with smaller clusters of natural killer (NK) cells, dendritic cells (DCs), FABP+ macrophages (FABP_M) and B cells. While cell counts corresponding to most of the clusters were largely similar in the two groups, cell counts for two, a neutrophil population that we named N3, and an IM population, identified as IM2, were 5.5× and 17.5× higher in cells isolated from LPS + PA14 mice compared to those from PA14 mice (Fig. 3a and Supplementary Table 1). Although we used enriched CD11b+ cells for scRNA-seq analysis, minor populations of CD11b− cells were identified as contaminants that included AMs and fibroblasts. Neutrophils were identified using a combination of marker genes. Expression of Asprv1, which was characterized as a neutrophil-specific marker in both mice and humans20,21,22, was useful to identify four neutrophil clusters N1-N4 (Fig. 3b, and Supplementary Fig. 2a, c). The expression of Ly6g, most commonly associated with mature neutrophils23, was variable in the neutrophil populations, as also observed in other studies24. Clusters N1-N4 also expressed Cxcr2, a key chemokine receptor expressed by both mouse and human neutrophils, and the S100 genes S100a8 and S100a9, also commonly expressed by neutrophils. N1 displayed features of a mature neutrophil population with high level of expression of Ly6g and Ngp. N3, which was much more abundant in the LPS + PA14 group, expressed higher levels of multiple ISGs compared to the other neutrophil clusters. N3 also displayed expression of major histocompatibility complex (MHC) genes. The three IM populations were distinguished based on differential expressions of the MHC genes, H2-Eb1 and H2-Ab1, and Lyve1, as described previously25,26,27. Although IM1s are described as Ly6C1−, the IM1s in our analysis expressed Ly6C, as was also noted in a study of pulmonary hypertension28. Conversely, having detected Lyve1 expression in a small fraction of the monocytes (Fig. 3b), we labeled the cells Ly6C+Lyve1lo. Figure 3c depicts the proportions of these cell clusters in the two groups of mice. The scRNA-seq data helped to localize the increased Aoah expression detected in the LPS + PA14 group in whole lung tissue (Fig. 1f), to N3 and two of the IM clusters, IM2 and IM3 (Fig. 3d).
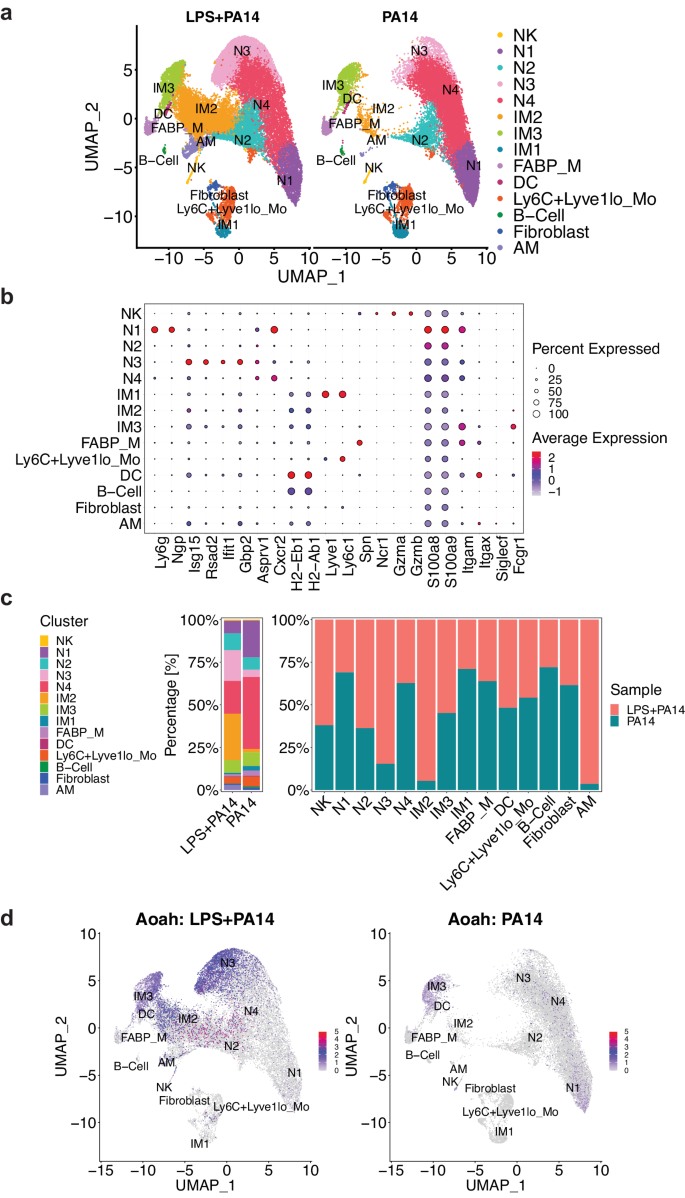
a UMAP embedding of the integrated expression profile of 46,826 cells from two conditions (LPS + PA14 and PA14), each comprised of three biological replicates. Distinct clusters are annotated and color-coded. b Expression of canonical markers used to annotate clusters, in conjunction with automated annotation using SingleR v2.2.0 (ImmGen reference scRNA-seq dataset). c Proportions of cell types represented in the two conditions, including the total count of cells in each cell type and the experimental condition. d Feature plot visualizing the expression of Aoah, the color of points (cells) represents the expression level of Aoah. Cell types are labeled, and the plot is split by experimental condition to highlight differences in expression levels between cells from LPS + PA14 and PA14 mice. n = 3 mice per group.
N3 and IM populations are distinguished by signatures of phagocytosis and cell killing
Differential expression analysis using the pseudobulk approach across aggregated cell clusters in the LPS + PA14 versus PA14 group was implemented using DESeq2. This analysis revealed increased expression of known LPS-inducible genes in the LPS + PA14 group, which included Saa3, an opsonin29, complement factor B (Cfb), which activates the alternate complement pathway and plays an important role in antibacterial defense30 and Aw112010, important in host defense and a suppressor of IL-10 gene expression31 (Fig. 4a). Upregulation of Aw112010 in the LPS + PA1 4 group was concordant with lower BAL IL-10 protein levels (Fig. 1g). Based on the differential expression analysis using single cell data, we examined DEGs across cell clusters in the two groups focusing on those clusters with the ability to phagocytose bacteria, which included the neutrophils, IMs and Ly6C+Lyve1loMos. As depicted in the volcano plots, some genes were upregulated in multiple clusters which included Saa3, Aw112010, the IFN-inducible gene, Gbp2, associated with bacterial killing32, and the chemokines Cxcl9 and Cxcl10, which in addition to their chemoattractant properties have potent antimicrobial functions33,34 (Supplementary Fig. 4). We examined the relative expression of selected genes important in bacterial clearance in all cell clusters in the two groups of mice, LPS + PA14 and PA14, presented as violin plots. These included genes related to opsonization, generation of reactive oxygen species (ROS) via the NADPH oxidase 2 (Nox) system, which is a central player in innate immunity effecting bacterial killing in neutrophils and macrophages35, Gbp2, Cxcl9, and Cxcl10. Expression of the genes, Saa3 and Cfb, was only detected in the LPS + PA14 group, the major signals being in the N3, IM2, and IM3 clusters (Fig. 4b). The expression profile of the Nox subunit genes was largely similar in the two groups. However, Cyba, which encodes p22phox, and associates with Cybb/gp91phox forming the membrane-bound subunits of the NADPH oxidase system35, was expressed at a higher level in N3 and the IM2 and IM3 populations in the LPS + PA14 mice relative to the PA14 mice. Gbp2 was robustly expressed in the LPS + PA14 mice, most prominently in the N3 and IM3 clusters, N2 also displaying some expression. Both Cxcl9 and Cxcl10 were found to be expressed more prominently in the LPS + PA14 mice, N3, IM2, and IM3 displaying the strongest signals among the neutrophil and IM populations, Cxcl10 being also expressed in the N2 cluster. Interestingly, although IFN-γ protein was detected by flow cytometry in the lung neutrophils, Ifng transcript was not detectable in any neutrophil population suggesting post-transcriptional regulation of IFN-γ production from very low levels of Ifng mRNA36.
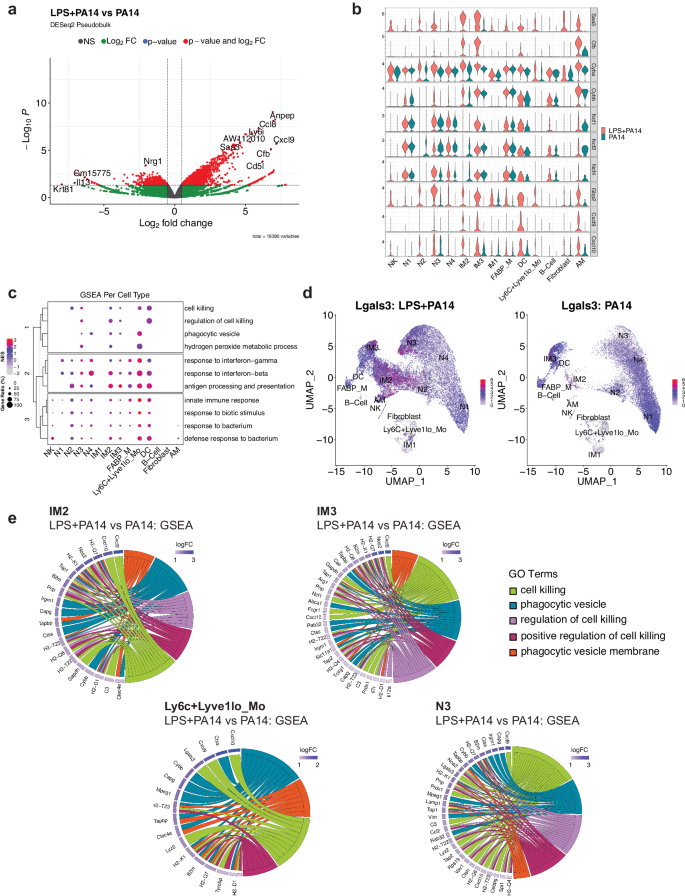
a Volcano plot of genes differentially expressed genes in the DESeq2-based pseudo-bulk analysis of LPS + PA14 versus PA14 cells. Red points indicate significance in both fold-change (log2FC > 0.5) and Benjamini-Hochberg (BH) adjusted p-value (p < 0.05). b Stacked violin plots highlighting select genes that were found to be differentially expressed between the two conditions (LPS + PA14 versus PA14). c A dotplot comparing the expression levels of core genes of select pathways of interest enriched in LPS + PA14 against PA14 cells. The size of the dot indicates the gene ratio, and the color of the dot shows the normalized enrichment score (NES). The dendrogram in the y-axis represents the clustering of the pathways by the NES. d Feature plot visualizing expression of Lgals3, and the color represents the average expression level across cell types. e Chord diagrams showing the pathways of interest from the GSEA. In each diagram, enriched clusters are shown on the right and genes contributing to the enrichment are shown on the left. Genes are colored by log fold-change value, and the color of the chords represent distinct pathway/ontology Terms.
To gain more biological insights underlying the N3, IM2 and IM3 cell types, we used Gene Set Enrichment Analysis (GSEA) to compare pathways activated/suppressed between LPS + PA14 mice versus the PA14 mice. We observed enrichment of various pathways in the LPS + PA14 mice that included response to both Type I and Type II IFN, response to bacteria together with pathways enriched in genes related to phagocytic vesicle/phagocytic vesicle membrane and cell killing (Supplementary Fig. 5). We further compared select pathways across all cell clusters, and the ones related to response to Type I and II IFNs and to bacteria were enriched in all cell clusters (Fig. 4c). However, the pathways related to cell killing and phagocytic vesicle showed selective enrichment in clusters N3, IM2, IM3 and Ly6C+Lyve1loMos. Also, only N3, IM2 and Ly6C+Lyve1loMos showed association with the GO term hydrogen peroxide metabolic process, which comprises genes such as Prdx5 and a superoxide dismutase (Sod) gene, Sod2, involved in reduction of oxidative burden in cells (Supplementary Data 1 includes GSEA data for all cell clusters). Although the neutrophil cluster N4 was more abundant in the PA14 group, it was enriched in genes associated with phagocytic vesicles but not cell killing. N2 showed a signal for cell killing but not phagocytic vesicle formation. None of these pathways was discernible in the mature neutrophil population N1 which was more numerous in the PA14 mice. The gene set cell killing in the cell cluster N3 included the genes Cxcl9, Cxcl10, and Lgals3, which encodes galectin-3 (Table S2). Prior studies have reported anti-bacterial functions of galectin-3 that include protection by galectin-3-expressing neutrophils from pneumococcal pneumonia37 and demonstration of direct bactericidal activity of galectin-338. Cell-intrinsic anti-bacterial function of galectin-3 was also described with its ability to direct the antimicrobial guanylate-binding protein, Gbp2, to pathogen-containing vesicles in macrophages37,38,39. Although mouse neutrophils are reported to express very low levels of galectin-3, unlike macrophages which constitute a rich source of the protein37, Fig. 4d shows Lgals3 expression in all neutrophil clusters and also in IM2, IM3, and the contaminating AMs in both groups of mice. However, with the greater abundance of N3 and IM2 in the LPS + PA14 mice compared to the PA14 mice, the former would be expected to produce higher levels of this antibacterial protein. Chord plot representation of the ontology terms shows the core genes enriched in these pathways and are involved in bacterial clearance via phagocytosis and killing mechanisms (Fig. 4e). For example, the phagocytic vesicle and phagocytic vesicle membrane GO terms include multiple genes related to antigen presentation, the gene Clec4e, known to promote phagocytosis40 and Cybb, the membrane-bound partner of Cyba (Fig. 4b). All of the cell clusters N3, IM2, IM3, and Ly6C+Lyve1loMos displayed the chemokine genes Cxcl10 and/or Cxcl9 associated with cell killing consistent with the known potent anti-bacterial properties of these chemokines33,34.
Pre-treatment with LPS in PA14-infected mice helps to educate neutrophils for phagocytosis and cell killing
Given that the scRNA-seq data suggested increased bacterial phagocytosis and killing in mice pre-exposed to LPS, we next performed functional experiments to validate the data. We focused on neutrophils for the phagocytosis experiments given their essential role in defense against P. aeruginosa12 and the impact of LPS in augmenting accumulation of neutrophils. To study phagocytosis, we performed intravital imaging using a reporter-GFP bacterium, PA-GFP, that we found elicited a similar outcome in mice as compared to PA14 with respect to physical appearance of the mice after infection, ensuing bacterial burden, and lung immune cell profile (Supplementary Fig. 6a, b). To assess phagocytosis of PA-GFP by neutrophils, mice were treated i.t. with either LPS or PBS followed in 3 days i.t. with PA-GFP. Quantitative fluorescence intravital lung microscopy (qFILM) was performed to assess phagocytosis in the lungs of live mice at 3–4 h post-infection. Neutrophils (red) were visualized in vivo using Pacific Blue (PB)-conjugated anti-Ly6G antibody (pseudocolored red) administered intravenously (i.v.) and the microcirculation was visualized by i.v. injection of Texas Red Dextran, pseudo-colored purple. As shown in Fig. 5a–e, and in Supplementary information, included as Supplementary movies 1 and 2, LPS treatment promoted phagocytosis of PA-GFP by the neutrophils, as evidenced by increased colocalization of PB and PA-GFP in the neutrophils (Fig. 5d). Also, the percentage of PB-eGFP colocalization was significantly higher in the LPS pre-exposed mice showing that colocalization was independent from the number of neutrophils per field of view (Fig. 5e). As observed in other experiments (Fig. 2b), neutrophil counts in the lungs of the LPS-treated mice were higher than those in the PA-GFP alone group (Fig. 5c). We examined expression of a gene set that was previously described to promote the phagocytotic function of neutrophils in all cell clusters41. The gene, Fcer1g, which encodes the Fcγ receptor, and plays an important role in phagocytosis and innate immunity42, was enriched in the N3 and IM2 clusters in the LPS + PA14 mice compared to that in the PA14 mice (Fig. 5f). Increased Rac1 expression in N3 was also detected in these mice, Rac1 being an important component of the plasma membrane Nox2 system in neutrophils35. Abca1 and Aif1, associated with efferocytosis of apoptotic cells and bacterial phagocytosis respectively43,44, were expressed at a higher level in IM3s in the LPS + PA14 mice compared to that in the PA14 mice.
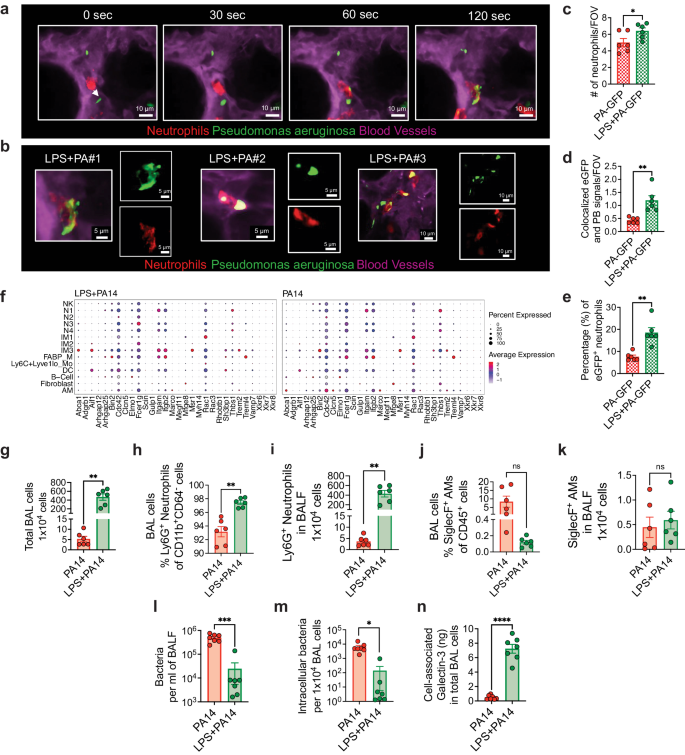
a Quantitative fluorescence intravital lung microscopy (qFILM) images of the same field of view (FOV) in the lung of an LPS + PA-GFP mouse at four different time points showing neutrophils crawling intravascularly (direction shown by white arrow) towards PA-GFP and finally phagocytosis of PA-GFP. Complete time series for (a) shown in Supplementary movies 1 and 2. b Magnified images showing phagocytosis in three different mice using merged channel pictures and isolated red and green channels. c Relative neutrophil counts in FOV in the two groups of mice. d qFILM data were analyzed as described in Methods. Phagocytosis of PA by neutrophils is shown as the average colocalization of eGFP and Pacific Blue (PB) signals within the FOV. e The percentage of eGFP+ neutrophils combining all fields of view. Representative images (a, b) and pooled data (c–e) from 3 independent experiments with n = 2 mice per group per experiment. Data analyzed using two-tailed unpaired t test with Welch's correction. f Dot plots showing the expression levels of phagocytosis-related genes in all cell types, separated by condition. Flow cytometry analysis of BAL cells (g) with neutrophils (h, i) comprising more than 90% of the BAL cells in PA14 and 98% in LPS + PA14 mice and AMs (j, k) comprising around 10% and 0.2% respectively. g–k n = 3 mice per group per experiment. Data pooled from 2 independent experiments and analyzed using two-tailed unpaired t test with Welch's correction. l Bacterial burden in BALF. m Intracellular bacterial load in BAL cells assessed by permeabilizing BAL cells. n Galectin-3 protein associated with BAL cells (primarily neutrophils). l–n Data pooled from 2 independent experiments with n = 7 mice per group. Data were log transformed using log base 10 to adjust for differences in standard deviation prior to analysis for (l, m). Data analyzed using two-tailed unpaired t test with Welch's correction. All data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data and exact P values are provided as a Source Data file.
We next collected BAL cells from the two groups of mice at 4 h post-infection to assess intracellular bacterial load having detected cell-killing signatures in the N3 cluster by GSEA. ~100-fold more BAL cells were recovered from the LPS + PA14 mice compared to the PA14 mice (Fig. 5g). The majority were neutrophils in both, the remaining cells being AMs (Fig. 5h-k). We have not yet interrogated the nature of the neutrophil subpopulations in the BALF. The BALF was plated for bacterial quantification while BAL cell pellets were permeabilized to assess intracellular bacterial burden. A significantly lower bacterial count was detected in the BALF of LPS + PA14 mice as compared to that in the PA14 mice, with a similar 2–3-log fold reduction in intracellular bacteria in the BAL cells of LPS + PA14 mice (Fig. 5l, m). It is important to note that since the differential bacterial counts were evident after permeabilization of the BAL cells, they reflected phagocytosed intracellular bacteria and not extracellular contaminants. Thus, while significantly higher numbers of neutrophils extravasated into the air spaces in the LPS + PA14 mice, they harbored few live bacteria. In contrast, the fewer neutrophils in the PA14 mice had a significantly higher load of live bacteria suggesting inability to efficiently kill the phagocytosed pathogen. Based on the GSEA data, we also assayed galectin-3 protein levels in the BAL cells and detected significantly higher expression associated with the cells from the LPS + PA14 group (Fig. 5n). Taken together, these results provided a functional correlate to the scRNA-seq data suggesting that neutrophils played a key role in phagocytosis and killing of bacteria in mice pre-exposed to LPS.
Galectin-3 levels in the lower respiratory tract of critically ill patients with acute respiratory failure correlate with neutrophil biomarkers and predict survival
Respiratory infections and their sequelae can precipitate acute respiratory failure (ARF). Given Lgals3 expression in neutrophils in the mouse data, its enrichment in the highly abundant N3 cluster in the LPS + PA14 mice associated with cell killing gene signature, and detection of significantly higher galectin-3 protein levels associated with BAL neutrophils in LPS + PA14 mice, we sought to determine whether clinically significant differences in galectin-3 levels may be present in the lower respiratory tract (LRT) of patients with ARF. We have recently demonstrated that host response biomarkers can be measured reproducibly in supernatant fluid of endotracheal aspirate (ETA) specimens obtained from mechanically ventilated patients with ARF, offering clinically valid discriminations for the type and etiology of ARF45. Therefore, we studied a cohort of 81 patients with severe ARF, either with acute respiratory distress syndrome (ARDS) per Berlin criteria (n<...
Comments
Post a Comment